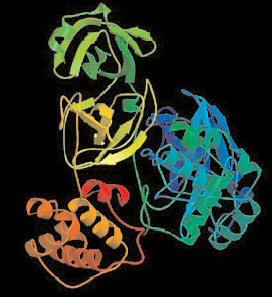
Using x-ray beams from the U.S. Department of Energy's (DOE's) Advanced Photon Source, Structural GenomiX, Inc., (SGX), a San Diego, California-based, structure-guided drug discovery company, has completed the three-dimensional crystal structure of the main protease from the Coronavirus that causes Severe Acute Respiratory Syndrome (SARS). The SGX Collaborative Access Team (SGX-CAT) beamline at sector 31 of the Advanced Photon Source, a national research facility funded by the DOE's Office of Science, Office of Basic Energy Sciences, and located at the DOE's Argonne National Laboratory, was used to collect data to solve the structure.
Recognizing the serious public health issue posed by SARS, SGX is making the crystal structure available to the research community by depositing it with the Protein Data Bank (PDB, www.rcsb.org), a public database accessed by researchers worldwide, in advance of publishing a paper in a refereed scientific journal, and is exploring collaborative opportunities to accelerate development of a treatment for the virus.
The determination was achieved via utilization of the SGX proprietary high-throughput gene-to-structure platform and its x-ray data collection facility at the Advanced Photon Source. SGX determined the high-resolution [1.86 .] crystal structure of the protease in a little over one month after receiving cDNA clones from the Genome Institute of Singapore (GIS). The scientists at GIS, with whom SGX is collaborating, were among the first to decode the genome of the SARS Coronavirus. They subsequently cloned the genes encoded by the viral genome and have made this information readily available to the scientific community worldwide.
The SARS main protease is widely believed to be the primary target for inhibitory antiviral agents, and the experimental determination of its crystal structure represents a major step forward in the discovery of new drugs to combat the virus. The three-dimensional crystal structure of the SARS Coronavirus main protease will enable structure-guided discovery of a drug that binds to the protease and inhibits the SARS virus replication. A similar strategy has succeeded previously with the human immunodeficiency virus protease for treatment of AIDS.
"SARS has been labeled the first '21st Century Epidemic'," said Joshua Lederberg, Ph.D., President Emeritus of The Rockefeller University, Nobel Laureate, and a leading expert in antiviral research. "There is no better example of the concert of effort, and the use of the most sophisticated insights and tools, than the leap from an enigmatic new syndrome to the solution of a high-resolution protein structure in a matter of a few months. The prompt sharing of this information by SGX bodes well for hopes of developing new drugs that may be our major armor if there is a recurrence of SARS."
"It is a testament to the power of the SGX technology platform that SGX has been able to produce this important crystal structure so rapidly, " said George Poste, D.V.M., Ph.D., Chairman of SGX. "This experimentally determined structure provides a highly accurate view of the three dimensional structure of the SARS main protease and also provides insights to facilitate drug discovery by allowing researchers to see in three dimensions precisely how inhibitors are binding to this important anti-SARS virus target."
Scientists have recently determined crystal structures of the main protease from other strains of human and porcine Coronaviruses that do not cause SARS. These data were used to calculate computer models of the structure of the SARS main protease. In contrast, SGX has determined an actual high-resolution experimental structure of the SARS main protease using x-ray diffraction. With the experimental structure and an ample supply of crystals of the SARS main protease in hand, SGX can now conduct iterative co-crystallization experiments of putative inhibitors with the SARS protease to guide discovery and optimization of inhibitors.
For more information on the Advanced Photon Source, visit us on the Web.
For more about Structural GenomiX, visit their Web site at https://stromix.com.
NOTE: See the New York Times online story at: NY Times