The original University of Texas at Austin “UT News” article by Nat Levy can be read here.
An enzyme variant created by engineers and scientists at The University of Texas at Austin (UT Austin) can break down environment-throttling plastics that typically take centuries to degrade in just a matter of hours to days. This discovery, which uses experimental results from studies at the U.S. Department of Energy’s Advanced Photon Source (APS) and is published in Nature, could help solve one of the world’s most pressing environmental problems: what to do with the billions of tons of plastic waste piling up in landfills and polluting our natural lands and water. The enzyme has the potential to supercharge recycling on a large scale that would allow major industries to reduce their environmental impact by recovering and reusing plastics at the molecular level.
“The possibilities are endless across industries to leverage this leading-edge recycling process,” said Hal Alper, professor in the McKetta Department of Chemical Engineering at UT Austin. “Beyond the obvious waste management industry, this also provides corporations from every sector the opportunity to take a lead in recycling their products. Through these more sustainable enzyme approaches, we can begin to envision a true circular plastics economy.”
The project focuses on polyethylene terephthalate (PET), a significant polymer found in most consumer packaging, including cookie containers, soda bottles, fruit and salad packaging, and certain fibers and textiles. It makes up 12% of all global waste.
The enzyme was able to complete a “circular process” of breaking down the plastic into smaller parts (depolymerization) and then chemically putting it back together (repolymerization). In some cases, these plastics can be fully broken down to monomers in as little as 24 hours.
Researchers at the Cockrell School of Engineering and College of Natural Sciences used a machine learning model to generate novel mutations to a natural enzyme called PETase that allows bacteria to degrade PET plastics. The model predicts which mutations in these enzymes would accomplish the goal of quickly depolymerizing post-consumer waste plastic at or near ambient temperatures, i.e., with little or no extra heating of the reaction mixture.
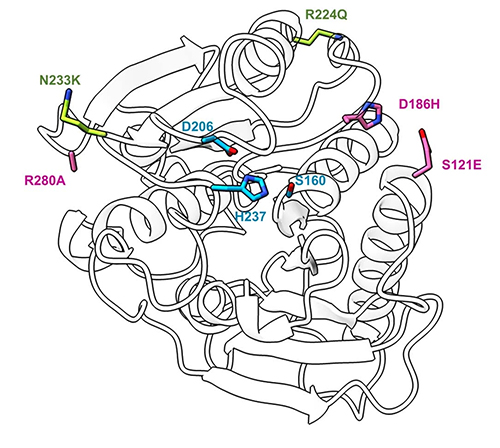
Through this process, which included studying 51 different post-consumer plastic containers, five different polyester fibers and fabrics and water bottles all made from PET, the researchers proved the effectiveness of the enzyme, which they are calling FAST-PETase (functional, active, stable and tolerant PETase). As part of the study, protein crystals of FAST-PETase were shipped from Texas to the Advanced Photon Source, an Office of Science user facility at Argonne National Laboratory, in Illinois. X-ray diffraction data were collected from individual crystals by graduate student Wantae Kim at home in Texas by using the remote-operation and user-friendly beamline capabilities of x-ray beamline 23-ID-B of the National Institute of General Medical Sciences and National Cancer Institute (GM/CA-XSD) structural biology facility at the APS (the APS is an Office of Science user facility at Argonne National Laboratory). Via an experimental technique called x-ray crystallography, these data (Fig. 1) enabled the researchers to determine the atomic structure of the enzyme in three dimensions, which provides insight into how FAST-PETase functions near ambient temperatures to break bonds in plastic.
“This work really demonstrates the power of bringing together different disciplines, from synthetic biology to chemical engineering to artificial intelligence,” said Andrew Ellington, professor in the Center for Systems and Synthetic Biology whose team led the development of the machine learning model.
Recycling is the most obvious way to cut down on plastic waste. But globally, less than 10% of all plastic has been recycled. The most common method for disposing of plastic, besides throwing it in a landfill, is to burn it, which is costly, energy intensive and spews noxious gas into the air. Other alternative industrial processes include very energy-intensive processes of glycolysis, pyrolysis, and/or methanolysis.
Biological solutions take much less energy. Research on enzymes for plastic recycling has advanced during the past 15 years. However, until now, no one had been able to figure out how to make enzymes that could operate efficiently at low temperatures to make them both portable and affordable at large industrial scale. FAST-PETase can perform the process at less than 50 degrees Celsius.
Up next, the team plans to work on scaling up enzyme production to prepare for industrial and environmental application. The researchers have filed a patent application for the technology and are eying several different uses. Cleaning up landfills and greening high waste-producing industries are the most obvious. But another key potential use is environmental remediation. The team is looking at a number of ways to get the enzymes out into the field to clean up polluted sites.
“When considering environmental cleanup applications, you need an enzyme that can work in the environment at ambient temperature. This requirement is where our tech has a huge advantage in the future,” Alper said.
(©The University of Texas at Austin 2022)
See: Hongyuan Lu1, Daniel J. Diaz1, Natalie J. Czarnecki1, Congzhi Zhu1, Wantae Kim1, Raghav Shroff1,2, Daniel J. Acosta1, Bradley R. Alexander1, Hannah O. Cole1, Yan Zhang1, Nathaniel A. Lynd1, Andrew D. Ellington1, and Hal S. Alper1*, “Machine learning-aided engineering of hydrolases for PET depolymerization,” Nature 604, 662 (28 April 2022). DOI: 10.1038/s41586-022-04599-z
Author affiliations: 1The University of Texas at Austin, 2DEVCOM ARL-South
Correspondence: * [email protected]
This work was financed under research agreement no. EM10480.26/ UTA16-000509 between the ExxonMobil Research and Engineering Company and The University of Texas at Austin. Sequencing was conducted at the Genomic Sequencing and Analysis Facility (RRID no. SCR_021713), SEM was conducted at the Microscopy and Imaging Facility (RRID no. SCR_021756) at the UT Austin Center for Biomedical Research Support, and AFM analysis was conducted at the Texas Materials Institute at UT Austin. N.A.L. and C.Z. thank the Welch Foundation for partial support of this research (Grant #F-1904). GM/CA-XSD has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). The crystallography study is supported by a grant from the National Institutes of Health (no. GM104896 to Y.J.Z.). The authors acknowledge the Texas Advanced Computing Center at The University of Texas at Austin for providing deep learning resources for neural network predictions and analysis that have contributed to the research results reported in this paper. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility, operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
The U.S. Department of Energy's APS at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
