Malaria is one of the most devastating diseases impacting human health, causing approximately half a million deaths each year. The disease is caused by infection with the intracellular parasite Plasmodium and is transmitted by female Anopheles mosquitoes during feeding. Of the six species of Plasmodium, P. falciparum is both the most lethal and responsible for most deadly cases. Some P. falciparum RIFINs—variant surface antigens expressed on infected red blood cells—bind to multiple inhibitory receptors, including LAIR1 and LILRB1. Researchers identified antibodies incorporating LILRB1 by screening plasma from donors from the Republic of Mali. B cell clones isolated from three of the donors showed large DNA insertions in the switch region that encodes LILRB1 extracellular domain 3 and 4 (D3D4) or D3 alone in the variable–constant (VH–CH1) elbow. By using a combination of biochemical and bioinformatic techniques, the researchers identified a large set of RIFINs that bound LILRB1 D3. Crystal and cryo-electron microscopy structures of RIFIN in complex with LILRB1 D3D4 revealed RIFIN–LILRB1 D3 interactions to be similar though distinct from those of RIFIN–LAIR1. The RIFIN–LILRB1 structures, including those obtained at the U.S. Department of Energy’s Advanced Photon Source and published in the journal Nature, further revealed an unconventional triangular architecture with the inserted LILRB1 domains opening the VH–CH1 elbow. Better understanding the mechanism that leads to receptor-binding specificity of RIFINs could guide the development of malaria vaccines and therapeutic antibodies.
The symptoms of malaria occur as Plasmodium parasites replicate within blood. This nutrient-rich environment also contains much of the host’s immune-defense machinery. To survive the immune response, Plasmodium parasites have evolved to replicate while hidden within host cells. Only a few functionally critical parasite proteins are exposed on host cell surfaces, which are grouped into large protein families, allowing a immune evasion strategy based on antigenic variation.
Repetitive interspersed families of polypeptides (RIFINs) are variable antigens expressed on infected red blood cells. They mediate formation of clusters of uninfected red blood cells surrounding a central P. falciparum-infected red blood cell that occurs in severe malaria. RIFINs bind LAIR1 and LILRB1—two inhibitory receptors found on natural killer cells, T cells, and B cells—playing a role in immune evasion by downregulating activation of these immune cells. RIFINs, therefore, may potentially be targets for protective immunity.
On the basis of their earlier discovery of LAIR1-containing antibodies in malaria-exposed individuals, researchers in the Antonio Lanzavecchia group at the Institute for Research in Biomedicine, Switzerland hypothesized that B cell clones with insertions of other inhibitory receptors recognized by P. falciparum, such as LILRB1, could be selected in the course of malaria infection. To test this, they screened 672 plasma samples from a cohort of donors from Mali and identified six individuals with LILRB1-containing antibodies. From three positive donors, the researchers isolated B cell clones that produced LILRB1-containing monoclonal antibodies bound to infected erythrocytes.
Through mass spectrometry and binding assays, the researchers identified seven RIFINs that were recognized by LILRB1-containing antibodies. Antibodies containing LILRB1 D3D4 or D3 inserts bound to the seven identified RIFINs, while deletion of the inserted domains completely abolished binding, indicating a critical role for D3. These RIFINs differ from those of an earlier report that bind to LILRB1 D1D2 domains, which compete for the nature ligand binding, suggesting distinct binding mechanisms.
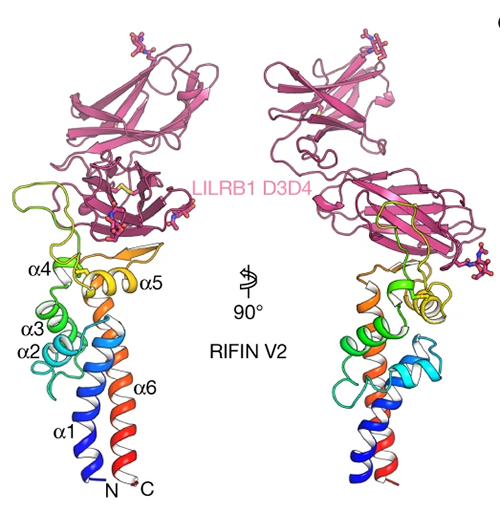
The researchers in the Peter Kwong group at the Vaccine Research Center, National Institutes of Health, then determined the cryo-electron microscopy structure of a RIFIN in complex with either LILRB1 D3D4 or a D3D4-containing Fab (antibody fragment) at 3.5-Å resolution. The structure revealed an unconventional triangular Fab architecture owing to the elbow insertion of LILRB1 D3D4 domains, with the VH–VL, D3D4, and CH1–CL domains composing the three apexes. In particular, the angle of the light chain elbow region was found to swing outward almost 90 degrees to accommodate the LILRB1 D3D4 insertion in the heavy chain elbow region.
Because the reconstruction of RIFIN in the LILRB1-RIFIN complex was not entirely clear, the researchers used x-ray crystallography data collected at the Southeast Regional Collaborative Access Team beamline 22-ID at the APS to determine the structure of the complex between the LILRB1 D3D4 domain and the RIFIN V2 domain to 2.6-Å resolution (Fig. 1). Together, the two structural papers of LILRB1–RIFIN complexes revealed that the V2 apex is the common binding site used by RIFINs to bind to different domains of the same receptor.
The results of this study provide insights into the role of RIFINs in immune evasion. The finding that phylogenetically distinct RIFINs target LILRB1 D1D2 and LILRB1 D3 provides a striking example of convergent evolution. Overall, the polygenicity and polymorphism of the RIFIN family is consistent with a strong selective pressure for evading the antibody response while developing different ways to bind to inhibitory receptors, a mechanism that can be effectively countered by receptor-based antibodies. ― Chris Palmer
See: Yiwei Chen1, 2, Kai Xu3, Luca Piccoli1, Mathilde Foglierini1,4, Joshua Tan1, Wenjie Jin1,2, Jason Gorman3, Yaroslav Tsybovsky5, Baoshan Zhang3, Boubacar Traore6, Chiara Silacci-Fregni1, Claudia Daubenberger7, Peter D. Crompton3, Roger Geiger1, Federica Sallusto1,2, Peter D. Kwong3, and Antonio Lanzavecchia1‡*, “Structural basis of malaria RIFIN binding by LILRB1-containing antibodies,” Nature 592, 639 (22 April 2021). DOI: 10.1038/s41586-021-03378-6
Author affiliations: 1Università della Svizzera italiana, 2ETH Zurich, 3National Institutes of Health, 4Swiss Institute of Bioinformatics (SIB), 5Frederick National Laboratory for Cancer Research, 6University of Sciences, Techniques and Technologies of Bamako, 7University of Basel ‡Present address: Humabs BioMed SA
Correspondence: * [email protected]
This work was partially supported by grants from the European Research Council (no. 670955 BROADimmune), the Fondation Louis-Jeantet, the Swiss Vaccine Research Institute, the Swiss National Science Foundation (grant no. 176165). The Mali study was funded by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was supported by the Intramural Research Program of the Vaccine Research Center, National Institution of Allergy and Infectious Diseases, National Institutes of Health (NIH), by the GenScript Innovation grant GS-IG-2018-003 (K.X.), and by federal funds from the Frederick National Laboratory for Cancer Research, NIH, under Contract HHSN261200800001 (Y.T.). The Southeast Region Collaborative Access team) is supported by its member institutions, and equipment grants (S10_RR25528, S10_RR028976 and S10_OD027000) from the National Institutes of Health. Some of this work was performed at the Simons Electron Microscopy Center, and/or the National Resource for Automated Molecular Microscopy, and/or the National Center for Cryo-EM Access and Training located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247) and NIH National Institute of General Medical Sciences (GM103310) with additional support from NYSTAR and the New York State Assembly Majority. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's Advanced Photon Source at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries, and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
