The original Brookhaven National Laboratory press release can be read here.
A team of scientists led by researchers from the U.S. Department of Energy’s (DOE’s) National Synchrotron Light Source II (NSLS II) have demonstrated a new technique for imaging proteins in 3-D with nanoscale resolution. Their work, based on studies at NSLS II and the DOE’s Advanced Photon Source (APS) and published in the Journal of the American Chemical Society, enables researchers to identify the precise location of proteins within individual cells, reaching the resolution of the cell membrane and the smallest subcellular organelles.
“In the structural biology world, scientists use techniques like x-ray crystallography and cryo-electron microscopy to learn about the precise structure of proteins and infer their functions, but we don’t learn where they function in a cell,” said corresponding author and NSLS-II scientist Lisa Miller. “If you’re studying a particular disease, you need to know if a protein is functioning in the wrong place or not at all.”
The new technique developed by Miller and her colleagues is similar in style to traditional methods of fluorescence microscopy in biology, in which a molecule called green fluorescent protein (GFP) can be attached to other proteins to reveal their location. When GFP is exposed to UV or visible light, it fluoresces a bright green color, illuminating an otherwise “invisible” protein in the cell.
“Using GFP, we can see if a protein is in subcellular structures that are hundreds of nanometers in size, like the nucleus or the cytoplasm,” Miller said, “but structures like a cell membrane, which is only seven to 10 nanometers in size, are difficult to see with visible light tags like GFP. To see structures the size of 10 nanometers in a cell, you benefit greatly from the use of x-rays.”
To overcome this challenge, researchers at NSLS-II teamed up with scientists at the Massachusetts Institute of Technology (MIT) and Boston University (BU) who developed an x-ray-sensitive tag called a lanthanide-binding tag (LBT). LBTs are very small proteins that can bind tightly to elements in the lanthanide series, such as erbium and europium.
“Unlike GFP, which fluoresces when exposed to UV or visible light, lanthanides fluoresce in the presence of x-rays,” said lead author Tiffany Victor, a research associate at NSLS-II. “And since lanthanides do not occur naturally in the cell, when we see them with the x-ray microscope, we know the location of our protein of interest.”
The researchers at NSLS-II, MIT, and BU worked together to combine LBT technology with x-ray-fluorescence. “Although LBTs have been used extensively over the last decade, they’ve never been used for x-ray fluorescence studies,” Miller said.
Beyond obtaining higher resolution images, x-ray fluorescence simultaneously provides chemical images on all trace elements in a cell, such as calcium, potassium, iron, copper, and zinc. In other studies, Miller’s team is researching how trace elements like copper are linked to neuron death in diseases like Alzheimer’s. Visualizing the location of these elements in relation to specific proteins will be key to new findings.
In addition to their compatibility with x-rays, LBTs are also beneficial for their relatively small size, compared to visible light tags.
“Imagine you had a tail attached to you that was the size of your whole body, or bigger,” Miller said. “There would be a lot of normal activities that you’d no longer be able to do. But if you only had to walk around with a tiny pig’s tail, you could still run, jump, and fit through doorways. GFP is like the big tail—it can be a real impediment to the function of a many proteins. But these little lanthanide-binding tags are almost invisible.”
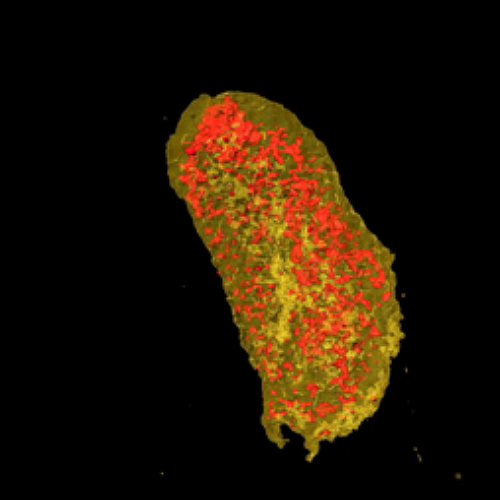
To demonstrate the use of LBTs for imaging proteins in 3-D with nanoscale resolution, the researchers at MIT and BU tagged two proteins in a bacterial cell—one cytoplasmic protein and one membrane protein. Then, Miller’s team studied the sample at the Hard X-ray Nanoprobe (HXN) beamline at NSLS-II and the Bionanoprobe at the Life Sciences Collaborative Access Team beamline 21-ID-D (now at X-ray Science Division 9-ID-B) at the APS — a DOE Office of Science user facility at the DOE’s Argonne National Laboratory.
“HXN offers the world-leading x-ray focus size, which goes down to about 12 nanometers. This was critical for imaging the bacterial cell in 3-D with nanoscale resolution,” said Yong Chu, lead beamline scientist at HXN. “We also developed a new way of mounting the cells on a specialized sample holder in order to optimize the efficiency of the measurements.”
By coupling the unparalleled resolution of HXN with the capabilities of LBTs, the team was able to image both of the tagged proteins. Visualizing the cell membrane protein proved LBTs can be seen at a high resolution, while imaging the cytoplasmic protein showed LBTs could also be visualized within the cell.
“At high concentrations, lanthanides are toxic to cells,” Victor said, “so it was important for us to show that we could treat cells with a very low lanthanide concentration that was nontoxic and substantial enough to make it past the cell membrane and image the proteins we wanted to see.”
Now, with this new technique demonstrated successfully, scientists hope to be able to use LBTs to image other proteins within the cell at a resolution of 10 nanometers.
See: Tiffany W. Victor1, Katherine H. O’Toole2, Lindsey M. Easthon2, Mingyuan Ge1, Randy J. Smith1, Xiaojing Huang1, Hanfei Yan1, Yong S. Chu1, Si Chen3, Doga Gursoy3, Martina Ralle4, Barbara Imperiali5, Karen N. Allen2, and Lisa M. Miller1,6*, “Lanthanide-Binding Tags for 3D X‑ray Imaging of Proteins in Cells at Nanoscale Resolution,” J. Am. Chem. Soc. 142, 2145 (2020). DOI: 10.1021/jacs.9b11571
Author affiliation: 1Brookhaven National Laboratory, 2Boston University, 3Argonne National Laboratory, 4Oregon Health Science University, 5Massachusetts Institute of Technology, 6Stony Brook University
Correspondence: *[email protected]
This work was supported by the United States Department of Energy (DOE) Office of Biological and Environmental Research (BER), as part of the “Environment Sensing and Response” Scientific Focus Area of the BER Genomic Science Program. T.W.V. was partially supported by the National Institutes of Health T32 Grant 5T32GM092714 and a Director’s Postdoctoral Fellowship at NSLS-II. B.I. and K.N.A. were supported by National Science Foundation MCB-1615252 and MCB-1614608, respectively. This research used beamline 3-ID (HXN) at NSLS-II, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
