The original University of Michigan Health System press release by Kara Gavin can be read here.
A banana a day may not keep the doctor away, but a substance originally found in bananas and carefully edited by scientists who carried out research at the U.S. Department of Energy’s Advanced Photon Source (APS) could someday fight off a wide range of viruses.
Bananas contain a substance that, when changed slightly by scientists, shows promise as an anti-virus drug. And the process used to create the virus-fighting form may help scientists develop even more drugs, by harnessing the “sugar code” that our cells use to communicate. That code gets hijacked by viruses and other invaders.
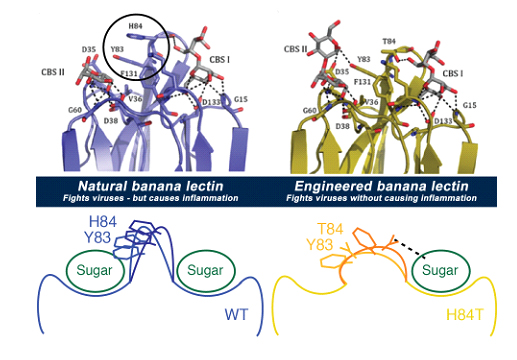
The new research focuses on a protein called banana lectin, or BanLec, that “reads” the sugars on the outside of both viruses and cells. Five years ago, scientists showed it could keep the virus that causes AIDS from getting into cells – but it also caused side effects that limited its potential use.
Now, in a new paper published in the journal Cell, an international team of scientists from the University of Michigan, the Ludwig-Maximilians-University Munich (Germany), the National University of Ireland, University College Dublin (Ireland), Université du Québec à Montrésal (Canada), Utah State University, Rockefeller University, Duke University, the University of Leuven (Belgium), and the University of North Carolina reports how they created a new form of BanLec that still fights viruses in mice, but doesn’t have a property that causes irritation and unwanted inflammation.
They succeeded in peeling apart these two functions by carefully studying the molecule in many ways, and pinpointing the tiny part that triggered side effects. Then, they engineered a new version of BanLec, called H84T, by slightly changing the gene that acts as the instruction manual for building it.
The result: a form of BanLec that worked against the viruses that cause AIDS, hepatitis C and influenza in tests in tissue and blood samples – without causing inflammation. The researchers also showed that H84T BanLec protected mice from getting infected by flu virus.
“What we’ve done is exciting because there is potential for BanLec to develop into a broad spectrum antiviral agent, something that is not clinically available to physicians and patients right now,” said David Markovitz, M.D., co-senior author of the new paper and a professor of internal medicine at the University of Michigan Medical School. “But it’s also exciting to have created it by engineering a lectin molecule for the first time, by understanding and then targeting the structure.”
The 26 scientists on the team worked together over several years to figure out exactly how BanLec worked against viruses, and then to build a better version. They were funded by the U.S. and European governments, and by foundations.
They used a wide range of scientific tools – including x-ray techniques employed by the U-M Center for Structural Biology working at the Life Sciences Collaborative Access Team x-ray beamline at the Argonne APS — that let them figure out the location of every atom in the original and new forms of BanLec.
Their efforts helped them understand how BanLec connects to both viruses and to sugar molecules on the outside of cells, and how it leads to irritation and other side effects by triggering signals that call in the “first responders” of the body’s immune system.
This understanding is what allowed them to change the gene in a way that fine-tuned the BanLec molecule. The new one still kept viruses out of cells, but doesn’t have the property that triggers the immune system response.
The new version of BanLec has one less tiny spot on its surface for sugars to attach, called a “Greek key” site. This makes it impossible for sugars on the surface of immune system cells called T cells to attach in multiple spots at once and trigger inflammation. But it still allows BanLec to grab on to sugars on the surface of viruses and keep them from getting into cells.
Several years of research still lie ahead before BanLec can be tested in humans. But Markovitz and his co-senior author Hashim M. Al-Hashimi, Ph.D., professor of biochemistry at Duke University and formerly professor of chemistry and biophysics at U-M, hope that their team’s work can help address the lack of antiviral drugs that work well against many viruses or against viruses that change rapidly, such as influenza.
“Better flu treatments are desperately needed,” said Markovitz, who treats patients with infectious diseases at the U-M Health System. “Tamiflu is only modestly effective, especially in critically ill patients, and influenza can develop resistance to it. But we also hope that BanLec could become useful in situations such as emergency pandemic response, and military settings, where the precise cause of an infection is unknown but a viral cause is suspected.”
The team continues to test H84T BanLec against other viruses in mice and tissue samples.
To see a brief animation of the BanLec molecule, click here.
See: Michael D. Swanson1,10, Daniel M. Boudreaux1, Loïc Salmon1, Jeetender Chugh1, Harry C. Winter1, Jennifer L. Meagher1, Sabine André2, Paul V. Murphy3, Stefan Oscarson4, René Roy5, Steven King1, Mark H. Kaplan1, Irwin J. Goldstein1, E. Bart Tarbet6, Brett L. Hurst6, Donald F. Smee6, Cynthia de la Fuente7, Hans-Heinrich Hoffmann7, Yi Xue8, Charles M. Rice7, Dominique Schols9, J. Victor Garcia10, Jeanne A. Stuckey1, Hans-Joachim Gabius2, Hashim M. Al-Hashimi1,8*, and David M. Markovitz1**, “Engineering a Therapeutic Lectin by Uncoupling Mitogenicity from Antiviral Activity,” Cell 163, 746 (October 22, 2015). DOI: 10.1016/j.cell.2015.09.056
Author affiliations: 1University of Michigan, 2Ludwig-Maximilians-University Munich, 3National University of Ireland, 4University College Dublin, 5Université du Québec à Montréal, 6Utah State University, 7Rockefeller University, 8Duke University, 9University of Leuven, 10University of North Carolina
Correspondence: *[email protected], **[email protected]
Work in the laboratories of D.M.M. and H.M.A-H. was supported by a National Institutes of Health (NIH) grant (1R01CA144043). H.-J.G. was supported by the ECfunded GlycoHIT program (contract no. 260600) and Training Network GLYCOPHARM (PITN-GA-2012-317297). M.D.S. and J.V.G. were supported by grants from the NIH (AI096138, AI073146, and P30 AI50410). P.V.M. has been supported by Marie Curie Intra-European Fellowships (500748, 514958, and 220948), the Programme for Research in Third-Level Institutions (PRTLI), administered by the Higher Education Authority, the Irish Research Council, Enterprise Ireland, and Science Foundation Ireland (04/BR/C0192, 06/RFP/CHO032, and 12/IA/1398). R.R. is grateful to the Natural Sciences and Engineering Research Council of Canada (NSERC) for financial support and for a Canadian Research Chair in Therapeutic Chemistry. The participation of A. Papadopoulos and T.C. Shiao is also acknowledged in the preparation of compounds 4–8. M.H.K. received support from the Concerned Parents for AIDS Research. D.S. was supported by KU Leuven grants (GOA 10/014 and PF 10/18), a European CHAARM grant (242135), and an equipment grant from the Fondation Dormeur, Vaduz. Work in the laboratory of C.M.R. was supported in part by PHS grants (R01 AI099284, R01 AI072613, and R01CA057973). Work at the Utah State University was supported by a grant (contract number HHSN2722010000391/HHSN27200005/A37) from the Respiratory Diseases Branch, Division of Microbiology and Infectious Diseases, NIAID, NIH. J.A.S., J.L.M, and H.M.A.-H. were partially supported by a grant (P50 GM103297) from the NIH. J.A.S. and J.L.M were also supported in part by the University of Michigan Center for Structural Biology. The Life Sciences Collaborative Access Team is supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). This research used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.