The original Purdue University press release by Emil Venere can be read here.
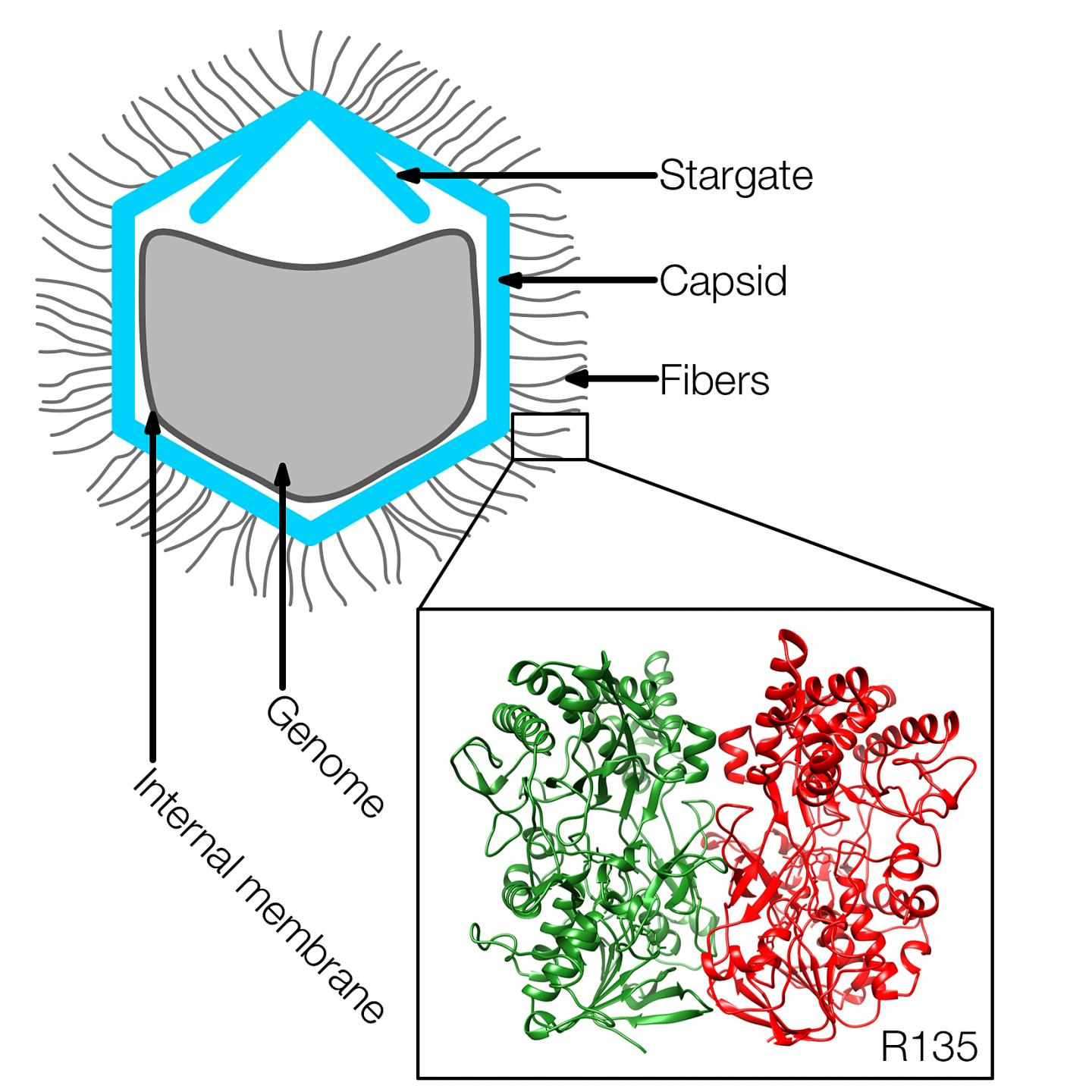
Researchers using high-brightness x-rays at the U.S. Department of Energy Office of Science’s Advanced Photon Source (APS) have discovered the structure of a key protein, R135, on the surface of an unusually large virus called the Mimivirus, aiding efforts to determine its hosts and unknown functions. Mimivirus is one of the largest viruses discovered to date, and little is known about how it infects its host. Determining the structure of R135, a protein that assists Mimivirus in entering its natural host, can provide new information about both the virus and its hosts. The results were published in the journal Structure.
The Mimivirus was initially thought to be a bacterium because it is much larger than most viruses. It was isolated by French scientists in 1992 but wasn't confirmed to be a virus until 2003.
In the laboratory, the virus has been studied while infecting amoebas, but its natural hosts in the wild and many details about the virus remain unknown, said Purdue University’s Michael Rossmann, lead author of the Structure article.
Using x-ray macromolecular crystallography from the National Institute of General Medical Sciences and National Cancer Institute (GM/CA-XSD) 23-ID-B and 23-ID-D beamlines at the Argonne APS, the team of researchers from Purdue discovered the structure of the enzyme-like protein called R135, which is contained in fibers on the outer surface of the virus. The structure of R135 is similar to an enzyme called aryl alcohol oxidase, which is found in a fungus and is involved in biodegrading lignin in plant cell walls.
“This could tell us something about the Mimivirus's natural hosts,” Rossmann said. “We think there must be another host, something different than amoebas and that this enzyme helps the virus get into this host. Perhaps R135 participates in the degradation of lignin so that the virus can enter a host such as lignin-containing algae.”
Also suggesting the possibility of alternative hosts is the recent discovery that Mimivirus has been found to be abundant in oysters. Antibodies against Mimivirus have been found in humans, and the virus has been discovered inside specialized cells in humans called macrophages, but whether it actually infects people is not known.
“I wouldn't say it infects humans, but it can propagate in humans because macrophages take up this virus and it can propagate in these,” said co-author Thomas Klose of Purdue. “Overall, there are many unknowns.”
Mimiviruses are among the largest known viruses. The virus approaches the size of bacteria and is about half of a micron in diameter, more than 10 times larger than the virus that causes the common cold and large enough to be seen with a light microscope. Other viruses are too small to be seen with conventional light microscopes.
“It's one of the biggest viruses and has about a thousand genes, many of which have unknown functions, and it seems to be like a melting pot of enzymes,” Klose said.
The R135 protein likely plays a key role in the virus's relationship with a smaller, “satellite virus” aptly called Sputnik. Sputnik is referred to as a satellite virus because it can only be propagated in combination with another virus, in this case the Mimivirus, which infects amoebas in laboratory research. While inside the single-cell amoeba, the DNA for both Sputnik and Mimivirus are replicated, allowing the viruses to multiply.
“It is very likely that this R135 is the protein that Sputnik uses to co-infect with Mimivirus,” Rossmann said.
See: Thomas Klose1, Dominik A. Herbst1‡ Hanyu Zhu2, Joann P. Max2, Hilkka I. Kenttämaa2, and Michael G. Rossmann1*, “A Mimivirus Enzyme that Participates in Viral Entry,” Structure 23, 1 (June 2, 2015). DOI: 10.1016/j.str.2015.03.023
Author affiliation: Purdue University ‡Present address: University of Basel
Correspondence: *[email protected]
This work was supported by the NIH (AI011219 to M.G.R.). The authors also gratefully acknowledge partial financial support by the Center for Direct Catalytic Conversion of Biomass to Biofuels (C3Bio), an Energy Frontier Research Center funded by the U.S. Department of Energy (DOE) Office of Science-Basic Energy Sciences under Award Number DE-SC0000997 (H.Z., H.I.K.). GM/CA-XSD has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.