Bispecific antibodies (BsAbs) are an emerging class of biological therapeutic agents designed to bind to two distinct regions on either the same or different targets, and therefore improve the body’s efficiency in fighting disease. However, their development for clinical use has been slowed, particularly due to issues with their structure that allow mispairing between the heavy and light chain regions of the antibody.
Researchers in this study designed a computer-assisted experimental approach, and using the Lilly Research Laboratories Collaborative Access Team beamline 31-ID-D at U.S. Department of Energy’s Advanced Photon Source (APS) to collect x-ray diffraction data, they produced novel mutant antibodies that were more stable and had the desired structure to reduce this indiscriminate pairing. These findings will be important in guiding future work to design new agents for varied therapeutic use.
Monoclonal antibodies (MAbs) have become an increasingly important source of therapeutic agents in recent years. These are antibodies that can recognize a single target antigen. However, BsAbs recognize two different antigens at the same time and can therefore bridge two distinct targets. This produces opportunities for increased therapeutic impact.
Although BsAbs have been produced for clinical use using a variety of approaches, production methods have been hindered by problems of manufacturability and issues with their physical and chemical properties.
Antibodies, or immunoglobulins, are large Y-shaped proteins used by the immune system to attack foreign substances such as bacteria, viruses, and even cancer cells. The two basic structural units of an antibody are heavy chains and light chains. The antibody recognizes a specific part (antigen) of the foreign substance – the tip of the “Y” contains a region that is specific for a particular part (epitope), on a target antigen, allowing them to bind together very precisely. The fragment antigen-binding (Fab fragment) is the region that binds to antigens, and contains one constant and one variable domain region of each of the heavy and light chain.
The “variable region” at the tip is made up of amino acids belonging to both light and heavy chains. This region also binds to antigen, but allows for variations in amino acid components, thereby enabling millions of antibodies with slightly different antigen-binding sites to exist. As a result of this, antibody heavy chains have evolved to bind light chains in a relatively nonspecific manner. This can be a major challenge with respect to generating BsAbs for clinical use, because this indiscriminate pairing leads to production of non-functional BsAbs. Specific pairing is essential for their optimal manufacturability and effectiveness.
Because Fab fragments can be produced in the laboratory, one solution to potentially produce a more specific BsAb and prevent mispairing of heavy and light chains involves the design of an antibody Fab with a fully orthogonal (perpendicularly arranged) structural interface where the heavy and light chain connect. Re-engineering a specific area of the antibody, known as the CH3 domain, plays a key role in efficient heavy chain pairing.
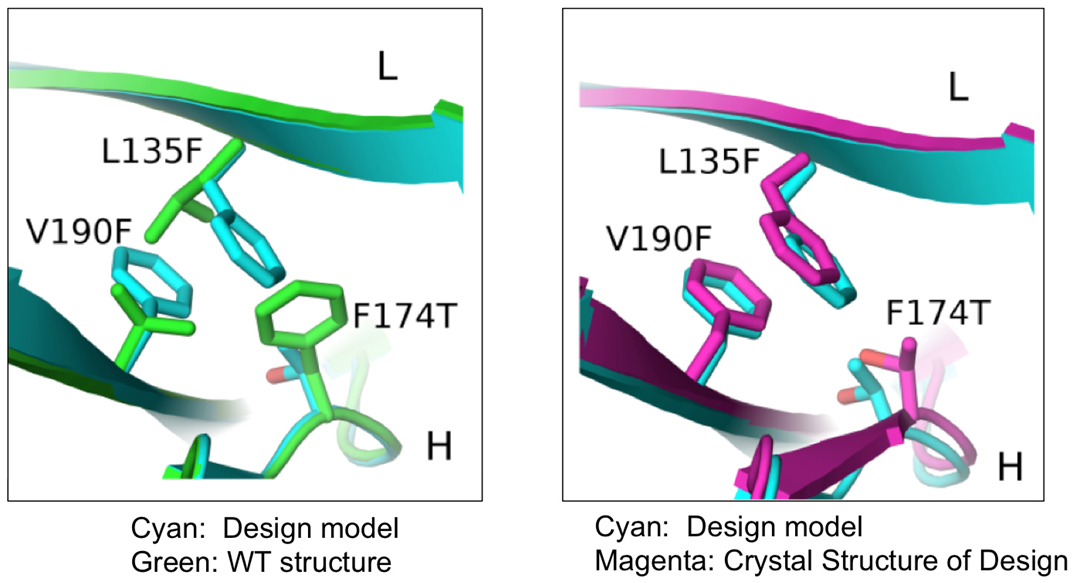
In this study, researchers from the Eli Lilly Biotechnology Center and the University of North Carolina at Chapel Hill used a combination of computational and rational design methods to produce novel BsAbs with the desired structure. This computer-aided approach involved choosing a suitable antibody to act as a protein “scaffold” from which new mutant Fabs were engineered following specific changes to the structure of the original scaffold. The researchers inserted mutations into the CH3 domains of various parental mAbs to allow expression of correctly paired BsAbs.
Using x-ray diffraction data sets collected from the 31-ID-D beamline at the at Argonne National Laboratory APS, an Office of Science user facility, they were then able to validate the three dimensional crystal structures of the newly produced proteins (Fig. 1).
As a result of this study, the researchers identified mutant antibodies that were more stable and prevented mispairing of heavy and light chains, facilitating expression of correctly assembled BsAbs using a variety of parental mAbs containing the mutant Fab interface.
These results will be important in guiding future work to investigate whether this strategy can be applied successfully to all antibodies, and may hold the potential to generate powerful therapeutic agents for the effective treatment of complex diseases.— Nicola Parry
See: Steven M. Lewis1, Xiufeng Wu2, Anna Pustilnik2, Arlene Sereno2, Flora Huang2, Heather L. Rick1, Gurkan Guntas1, Andrew Leaver-Fay1, Eric M. Smith2, Carolyn Ho2, Christophe Hansen-Estruch2, Aaron K. Chamberlain2, Stephanie M. Truhlar2, Elaine M. Conner1, Shane Atwell2, Brian Kuhlman1*, and Stephen J. Demarest2**, “Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface,” Nat. Biotechnol. 32(2), 191 (February 2014). DOI: 10.1038/nbt.2797
Author affiliations: 1University of North Carolina at Chapel Hill, 2Eli Lilly Biotechnology Center
Correspondence: * [email protected], ** [email protected]
This work was supported by the Lilly Research Laboratories and the Lilly Research Award Program. Use of the Lilly Research Laboratories Collaborative Access Team beamline was provided by Eli Lilly Company, which operates the facility. This research used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Argonne National Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov