When SARS-CoV-2, the virus that causes COVID-19, enters a person’s cells, it hijacks those cells to make more viruses. First SARS-CoV-2 releases its RNA into the host cell. Then the host ribosomes translate the viral RNA into two giant protein chains (polyproteins). One protein in the giant chain, called MPro, cleaves the chain into smaller proteins, which help create more viruses and, therefore, more infection. Because of MPro’s role in initiating the viral replication process, the protein has become a target for antiviral drug developers. Recently, a team of scientists using high-brightness x-rays at the U.S. Department of Energy’s Advanced Photon Source (APS) has determined x-ray crystallographic structures of MPro cleaving the polyprotein at ten cleavage sites. Their findings, published in the journal Nature Communications, provide information about the mechanistic steps and molecular interactions that initiate viral replication, which can be used to inform antiviral therapeutic development for COVID-19, as well as other conditions for which MPro may be responsible.
Viruses can’t reproduce on their own; they need a human or animal cell to make other viruses and continue their infectious rampage. The SARS-CoV-2 virus, which causes COVID-19, employs its spike protein to enter a human cell. Once inside, the virus’s protective coating dissolves, and it dumps its genetic material—RNA—into the host cell. This RNA contains all the instructions the virus needs to replicate. What’s more, it comes in a handy form that is ready for a human cell to translate into proteins that will compose the next generation of viruses.
The SARS-CoV-2 RNA includes instructions for four proteins that make up the virus’s structure—its spike protein, protective coating, and the like—and sixteen proteins that replicate the virus. The replication process begins when the host’s ribosomes translate the replication genes into two gigantic protein chains called polyproteins.
Before replication can continue, however, these gigantic chains must be chopped up into their constituent proteins. Remarkably, the molecule that does the chopping is itself contained in the polyprotein and must hack its way out of the chain before attending to its neighbors.
The chopping molecule is called MPro. The team of scientists responsible for the current work previously captured crystal structures of MPro chopping its way out of the polyprotein by recognizing its C terminal tail and cleaving itself from it. For this work, they mutated the C terminal tail of the MPro gene to the genetic sequences adjacent to the ten cleavage sites, tricking MPro into recognizing itself at the other cleavage sites. It took many tries—over 500—but eventually they were able to capture crystals of the same sort of complexes with the ten cleavage sites that they had previously captured of MPro cleaving itself from its own C terminal tail.
The scientists sent crystals that showed electron density for the bound sequences to three different facilities. Diffraction data at 100 K were collected at the National Institute of General Medical Sciences and National Cancer Institute (GM/CA-XSD) x-ray beamlines 23-ID-B and 23-ID-D at the APS, an Office of Science user facility at Argonne National Laboratory, using JBlueIce for data collection; beamlines CMCF-BM at the Canadian Light Source using MxDC for data collection; and beamlines 5.0.1 and 5.0.2 at the Advanced Light Source at Lawrence Berkeley National Laboratory using b4 for data collection.
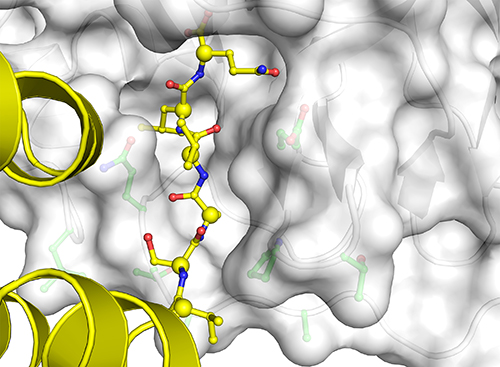
Now they were able to observe all the different conformations and catalytic features that accompanied MPro’s binding these multiple substrates: specifically, they observed the structure of the six amino acids that precede each cleavage site and contribute to the atomic interactions that determine how MPro binds the proteins; the different conformations the binding pockets undertake to adjust for the different sequences that they bind; and different approaches the substrates take in getting into MPro’s binding groove (Fig. 1).
But viruses are very tricky, and recent research has shown that MPro cleaves not only its own viral proteins but also host proteins in lung and kidney cells, enabling the virus to evade the human immune system and contributing to the virus’s pathogenicity.
Therefore, MPro has become a major focus for the development of a drug known as a direct-acting antiviral (DAA), which mimics substrate binding to MPro’s active site. The detailed information on substrate recognition reported in this research presents valuable insight into how to target the active site structure and mechanistic features of MPro’s essential role in viral replication and its ability to attack human targets. ― Judy Myers
See: Jaeyong Lee1,2, Calem Kenward1, Liam J. Worrall1, Marija Vuckovic1, Francesco Gentile1, Anh-Tien Ton1, Myles Ng1, Artem Cherkasov1, Natalie C. J. Strynadka1*, and Mark Paetzel2**, “X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation,” Nat. Commun. 13:5196 (2022). DOI: 10.1038/s41467-022-32854-4
Author affiliations: 1The University of British Columbia,2Simon Fraser University
Correspondence: * ncjs@mail.ubc.ca; ** mpaetzel@sfu.ca
This work was funded by operating grants from the Canadian Institutes of Health Research to N.C.J.S., M.P., and A.C.. N.C.J.S. and A.C. are Tier I Canada Research Chairs. C.K. is funded through a UBC Killam Doctoral Scholarship at UBC.We thank GM/CA-XSD beamline staff at beamline 23-ID-B at the APS for access and support. GM/CA-APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289. We thank staff at beamline 5.0.1 of the Advanced Light Source, a U.S. Department of Energy (DOE) Office of Science user facility under Contract No. DE-AC02-05CH11231, is supported in part by the ALS-ENABLE program funded by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01. Part or all of the research described in this paper was performed using beamline CMCF-BM at the Canadian Light Source, a national research facility of the University of Saskatchewan, which is supported by the Canada Foundation for Innovation (CFI), the Natural Sciences and Engineering Research Council (NSERC), the National Research Council (NRC), the Canadian Institutes of Health Research (CIHR), the Government of Saskatchewan, and the University of Saskatchewan. This research used resources of the Advanced Photon Source, a U.S. DOE Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The U.S. Department of Energy's APS at Argonne National Laboratory is one of the world’s most productive x-ray light source facilities. Each year, the APS provides high-brightness x-ray beams to a diverse community of more than 5,000 researchers in materials science, chemistry, condensed matter physics, the life and environmental sciences, and applied research. Researchers using the APS produce over 2,000 publications each year detailing impactful discoveries and solve more vital biological protein structures than users of any other x-ray light source research facility. APS x-rays are ideally suited for explorations of materials and biological structures; elemental distribution; chemical, magnetic, electronic states; and a wide range of technologically important engineering systems from batteries to fuel injector sprays, all of which are the foundations of our nation’s economic, technological, and physical well-being.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC, for the U.S. DOE Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.
